DNA profiles vary from person to person.
When profiles from a single VNTR locus from unrelated individuals are compared, the profiles are normally different.
However it is POSSIBLE for 2 individuals to have the same profile are 1 - 2 loci
The chance of more than 1 person having the same DNA profile at 4, 5 or 6 different VNTR loci is extremely small.
So what is DNA Profiling and how can we make it a reliable piece of physical evidence in court?
Each of us has a unique DNA profile or fingerprint. A technique called electrophoresis is used to obtain DNA profiles, relying on sections of our DNA that are known as non-coding DNA (DNA that does not code for a protein). To make it a reliable evidence, forensic scientists can obtain different sets of DNA and run electrophoresis on it, analysing the number of tandem repeats and the number of bands which are identical between the suspect and the DNA evidence.
How can DNA be used to identify an individual?
Almost every cell in our bodies contains DNA, the genetic material that programs how cells work. 99.9 percent of human DNA is the same in everyone, meaning that only 0.1 percent of our DNA is unique!
Each human cell contains three billion DNA base pairs. Our unique DNA, 0.1 percent of 3 billion, amounts to 3 million base pairs. This is more than enough to provide profiles that accurately identify a person.
What is DNA Profiling?
Living organisms that look different or have different characteristics (traits) also have different DNA sequences. The more varied the organisms, the more varied the DNA sequences.
DNA fingerprinting is a very quick way to compare the DNA sequences of any two living organisms or more,

How to make a DNA Profile

1. Isolation of DNA
2. Cutting, sizing, and sorting
3. Visualizing the DNA fingerprint
a. Transfer of DNA to nylon
b. Probing
c. DNA profile
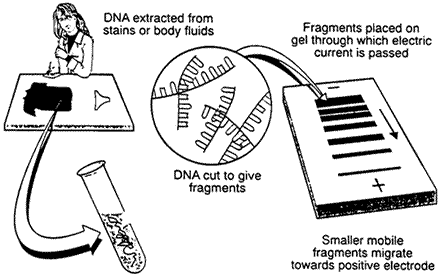
1.Isolation of DNA
DNA must be recovered from the cells or tissues of the body. Only a small amount of tissue - like blood, hair, or skin - is needed. For example, the amount of DNA found at the root of one hair is usually sufficient.
2.Cutting, sizing, and sorting

Special enzymes called restriction enzymes are used to cut the DNA at specific places.
For example, an enzyme called EcoR1, found in bacteria, will cut DNA only when the sequence GAATTC occurs.

Sizing & Sorting
The DNA pieces are sorted according to size by a sieving technique called electrophoresis.
The DNA pieces are passed through a gel made from seaweed agarose (a jelly-like product made from seaweed). This technique is the biotechnology equivalent of screening sand through progressively finer mesh screens to determine particle sizes.

Is this similar to the chromatography that you have done earlier?
How is it similar and different?
Yes. It is similar to chromatography. Like chromatography, the difference in results is based on the solvent's molecular weight. However, they are different as chromatography is conducted on paper whereas Electrophoresis is conducted on agar for the solvent to move more easily and cover a greater distance.
3. Visualizing the DNA fingerprint
a. Transfer of DNA to nylon
The stained fragments of DNA do not last long in the soft gel. Investigator will carry out the next phase to “fix” the DNA fragment permanently for further investigation.
The distribution of DNA pieces is transferred to a nylon sheet by placing the sheet on the gel and soaking them overnight.
b. Probing
ADDING RADIOACTIVE PROBES Adding radioactive or colored probes to the nylon sheet produces a pattern called the DNA fingerprint. Each probe typically sticks in only one or two specific places on the nylon sheet.
REMOVE PROBES Excess probes are washed away leaving behind the unique DNA bands.
c. DNA profile
The final DNA fingerprint is built by using several probes (5-10 or more) simultaneously.
The radioactive DNA pattern is transferred to X-ray film by direct exposure.
The resultant visible pattern after development is the DNA FINGERPRINT
No comments:
Post a Comment